| Bacterial Flagella: The Proton Turbine Model |
Here we will do some thought experiments with the proton turbine model of bacterial flagella rotation. It is well established
experimentally that rotation of the bacterial flagella motor requires a flow of protons across the cytoplasmic membrane (the
inner membrane of Gram negatives) and that these protons (essentially positive electricity) flow through the MotA/B of the
stator ring which consists of a number of subunits which functions as proton channels. The exact number varies with
species and the stator ring has been variously described as having 11, 13 or 16-fold symmetry. Evidence suggests that
these subunits may form something like 8 functional torque generators. The rotor (MS ring) also comprises 16 subunits
and accelerates in 16 discrete steps when starting up (when the motor is artificially switched off then on). Thus, we have
adopted a model in which there are 8 proton channels surrounding an M-ring with 16 subunits of alternating electric
charge, as shown below.
experimentally that rotation of the bacterial flagella motor requires a flow of protons across the cytoplasmic membrane (the
inner membrane of Gram negatives) and that these protons (essentially positive electricity) flow through the MotA/B of the
stator ring which consists of a number of subunits which functions as proton channels. The exact number varies with
species and the stator ring has been variously described as having 11, 13 or 16-fold symmetry. Evidence suggests that
these subunits may form something like 8 functional torque generators. The rotor (MS ring) also comprises 16 subunits
and accelerates in 16 discrete steps when starting up (when the motor is artificially switched off then on). Thus, we have
adopted a model in which there are 8 proton channels surrounding an M-ring with 16 subunits of alternating electric
charge, as shown below.
In this model, one proton is always assumed to occupy each Mot channel. Evidence suggests that some 50 protons flow
through each channel during each revolution of the rotor. The rate of flow through the channel does not enter into our
model, we simply assume that at any one instant there is one proton in each channel. Clearly, more charges could be
present at any one instant which will effect the forces exerted. The Mot channel might also partially screen the enclosed
charge, but we assume that the rotor sees a single positive charge in each Mot channel at any instant. Experimental
evidence suggests that each Mot unit is exerting force on the rotor most of the time, so the assumption of constant
occupancy seems a reasonable one. As seen in the plan view of the motor above, the presence of the protons will cause
the rotor to rotate counter-clockwise (CCW) one notch. To maintain the rotation we assume staggered lines of alternating
charge as shown in Fig. 1. We can then imagine a series of steps as the proton moves along the channel, which reset the
rotor ready for the next proton to enter. How many steps there are is not important for our calculations here, but the
concept of the model is illustrated in Figs. 1 and 3.
through each channel during each revolution of the rotor. The rate of flow through the channel does not enter into our
model, we simply assume that at any one instant there is one proton in each channel. Clearly, more charges could be
present at any one instant which will effect the forces exerted. The Mot channel might also partially screen the enclosed
charge, but we assume that the rotor sees a single positive charge in each Mot channel at any instant. Experimental
evidence suggests that each Mot unit is exerting force on the rotor most of the time, so the assumption of constant
occupancy seems a reasonable one. As seen in the plan view of the motor above, the presence of the protons will cause
the rotor to rotate counter-clockwise (CCW) one notch. To maintain the rotation we assume staggered lines of alternating
charge as shown in Fig. 1. We can then imagine a series of steps as the proton moves along the channel, which reset the
rotor ready for the next proton to enter. How many steps there are is not important for our calculations here, but the
concept of the model is illustrated in Figs. 1 and 3.
Figure 2.
Figure 3a, left.
The rotor advances one notch (to the
right) due to electrostatic attraction. An
odd number of steps fails to reset the
rotor if we assume the departing photon
effects a rotation. (It might not affect a
rotation as it may repel the incoming
positive charge as it exits).
Figure 3b, below.
An even number of steps resets the rotor
(there are arguments which can
conceivably reset the rotor after an odd
number of steps, but our simplistic
argument illustrates the essence of the
model).
The rotor advances one notch (to the
right) due to electrostatic attraction. An
odd number of steps fails to reset the
rotor if we assume the departing photon
effects a rotation. (It might not affect a
rotation as it may repel the incoming
positive charge as it exits).
Figure 3b, below.
An even number of steps resets the rotor
(there are arguments which can
conceivably reset the rotor after an odd
number of steps, but our simplistic
argument illustrates the essence of the
model).
Figure 4. Geometry of one sector of the model. O is the centre of the rotor (M ring) of
radius r. A proton is in the middle of the adjoining Mot complex which has a radius a. We
need to find the electrostatic Coulomb force of attraction between the negative charge on
the rotor and the proton (which acts towards the H+ along c). There is an equivalent
repulsive force between the positive site and the H+. Forces with more remote charge
sites on the M ring are ignored in this first approximation (but will serve to reduce the net
force slightly). Taking the radius of the M ring as 20 nm and the radius of the Mot as 1
nm, we can find the charge separation, c, as shown below:
radius r. A proton is in the middle of the adjoining Mot complex which has a radius a. We
need to find the electrostatic Coulomb force of attraction between the negative charge on
the rotor and the proton (which acts towards the H+ along c). There is an equivalent
repulsive force between the positive site and the H+. Forces with more remote charge
sites on the M ring are ignored in this first approximation (but will serve to reduce the net
force slightly). Taking the radius of the M ring as 20 nm and the radius of the Mot as 1
nm, we can find the charge separation, c, as shown below:
Calculation of Model
Figure 1. The proton turbine model of flagella rotation assumes that the driving
force for the flagella rotor is electrostatic and due principally to the flow of
protons through the stator ring.
force for the flagella rotor is electrostatic and due principally to the flow of
protons through the stator ring.
Figure 5. The calculation of charge separation c. This separation is used to calculate the
Coulomb force of electrostatic attraction (assuming no screening or unit charges).
Coulomb force of electrostatic attraction (assuming no screening or unit charges).
Figure 6. Calculation of the electrostatic attractive force. We are
interested in the component of this force tangential to the
circumference of the rotor (the radial component contributes no
torque). To resolve the force vector we calculate angle alpha below.
interested in the component of this force tangential to the
circumference of the rotor (the radial component contributes no
torque). To resolve the force vector we calculate angle alpha below.
Figure 7. Calculation of the angle alpha. This is used to derive the
tangential component of the electrostatic force below.
tangential component of the electrostatic force below.
Figure 8. Calculation of the total tangential force which turns the rotor. The attractive force is
multiplied by 2, since there is also a repulsive force which helps turn the rotor CCW and this is
due to the topmost charge in Fig. 4 being repelled by the proton. By symmetry, the tangential
component of this force has equal direction and magnitude to the attractive force. The total
force is further multiplied by 8, to account for the eight Mot units, resulting in the total tangential
force which turns the rotor. To apply this force we need to know the masses being acted upon.
multiplied by 2, since there is also a repulsive force which helps turn the rotor CCW and this is
due to the topmost charge in Fig. 4 being repelled by the proton. By symmetry, the tangential
component of this force has equal direction and magnitude to the attractive force. The total
force is further multiplied by 8, to account for the eight Mot units, resulting in the total tangential
force which turns the rotor. To apply this force we need to know the masses being acted upon.
Figure 9. Masses of the flagellum and its motor, obtained from the literature on the
basis of molecular mass and copy number of each protein component. This is used to
calculate the acceleration of the flagellum from Newton's Second Law, as shown below.
basis of molecular mass and copy number of each protein component. This is used to
calculate the acceleration of the flagellum from Newton's Second Law, as shown below.
Figure 10. The angular speed of the flagellum after 1s of acceleration by the Coulomb force.
The rotor can not accelerate indefinitely since resistive forces increase as the rotor speeds up
and this eventually balances the accelerating force. According to the literature, bacteria
decelerate and accelerate very rapidly, and assuming one second to reach maximum speed
seems reasonable for a loaded flagellum. However, this calculation gives far too high a value.
Bacteria flagella rotate at a maximum speed of 200-800 Hz when bearing no load other than
that due to the surrounding water and internal friction. In fact, with no additional loadings or
drag at all our motor would rapidly encounter relativistic effects by moving at a significant
fraction of the speed of light! In reality the flagellum can not accelerate without limit because
drag and frictional forces oppose its motion and these forces grow larger as the flagellum
accelerates until the two are balanced and then no further acceleration is possible.
The rotor can not accelerate indefinitely since resistive forces increase as the rotor speeds up
and this eventually balances the accelerating force. According to the literature, bacteria
decelerate and accelerate very rapidly, and assuming one second to reach maximum speed
seems reasonable for a loaded flagellum. However, this calculation gives far too high a value.
Bacteria flagella rotate at a maximum speed of 200-800 Hz when bearing no load other than
that due to the surrounding water and internal friction. In fact, with no additional loadings or
drag at all our motor would rapidly encounter relativistic effects by moving at a significant
fraction of the speed of light! In reality the flagellum can not accelerate without limit because
drag and frictional forces oppose its motion and these forces grow larger as the flagellum
accelerates until the two are balanced and then no further acceleration is possible.
Figure 12. For every torque there is an equal and opposite torque (Newton's third Law of
Motion) and the as the rotor drives flagellum rotation CCW so a force of equal magnitude
drives rotation of the cell body CW. Observations suggest that the cell body rotates about 10
rps, which our model correctly predicts within an order of magnitude.
Motion) and the as the rotor drives flagellum rotation CCW so a force of equal magnitude
drives rotation of the cell body CW. Observations suggest that the cell body rotates about 10
rps, which our model correctly predicts within an order of magnitude.
Considering the mass of water displaced by the flagellum in one second (by assuming one turn of the helix
every 2 micrometres, i.e. a filament wavelength of two micrometres) and a speed of 100 Hz we obtain an
estimate of the volume of water displaced, and with a low Re (creeping flow) this should be a good estimate
of the momentum of the displaced water directed behind the cell and by conservation of momentum we can
obtain the forward velocity of the cell. That is: we can calculate the linear momentum of the displaced water
and by applying conservation of linear momentum work out the expected speed of the swimming bacterium.
For a bacterial cell of 2 cubic micrometres (a typical Escherichia coli cell 2 micrometres long and one
micrometre wide), and a rotation speed of 100 Hz (a typical working speed) we obtain a predicted speed
(ignoring translational drag) of 32 micrometres / s. This is close to the expected 20 micrometres / s.
Conclusion
We have worked through a calculation of the torques produced by a proton turbine model and found that
electrostatic forces produce torques of the right magnitude. It is well-established that protons flow through the
Mot complex and that this drives flagella rotation. However, we still do not know whether this is due to simple
electrostatic attraction or whether the Mot complex undergoes some kind of conformational change (such as
extending a charged arm to bind reversibly to the rotor). Many molecular motors do work by conformational
changes, though this ultimately involves electrostatic and/or hydrophobic interactions to effect binding.
However, what calculations like the one presented here demonstrate is that electrostatic forces alone should
be sufficiently strong.
Article last updated: 14th Sept. 2014.
every 2 micrometres, i.e. a filament wavelength of two micrometres) and a speed of 100 Hz we obtain an
estimate of the volume of water displaced, and with a low Re (creeping flow) this should be a good estimate
of the momentum of the displaced water directed behind the cell and by conservation of momentum we can
obtain the forward velocity of the cell. That is: we can calculate the linear momentum of the displaced water
and by applying conservation of linear momentum work out the expected speed of the swimming bacterium.
For a bacterial cell of 2 cubic micrometres (a typical Escherichia coli cell 2 micrometres long and one
micrometre wide), and a rotation speed of 100 Hz (a typical working speed) we obtain a predicted speed
(ignoring translational drag) of 32 micrometres / s. This is close to the expected 20 micrometres / s.
Conclusion
We have worked through a calculation of the torques produced by a proton turbine model and found that
electrostatic forces produce torques of the right magnitude. It is well-established that protons flow through the
Mot complex and that this drives flagella rotation. However, we still do not know whether this is due to simple
electrostatic attraction or whether the Mot complex undergoes some kind of conformational change (such as
extending a charged arm to bind reversibly to the rotor). Many molecular motors do work by conformational
changes, though this ultimately involves electrostatic and/or hydrophobic interactions to effect binding.
However, what calculations like the one presented here demonstrate is that electrostatic forces alone should
be sufficiently strong.
Article last updated: 14th Sept. 2014.
Thus when we consider drag forces, our model gives a sensible prediction. There are also internal frictional
forces which we have ignored, however, experiments have shown that in the absence of the filament, the
rotor will spin at about 200 000 rps, suggesting that internal friction is negligible compared to the drag on the
flagellum filament which normally rotates at about 200 Hz.
forces which we have ignored, however, experiments have shown that in the absence of the filament, the
rotor will spin at about 200 000 rps, suggesting that internal friction is negligible compared to the drag on the
flagellum filament which normally rotates at about 200 Hz.
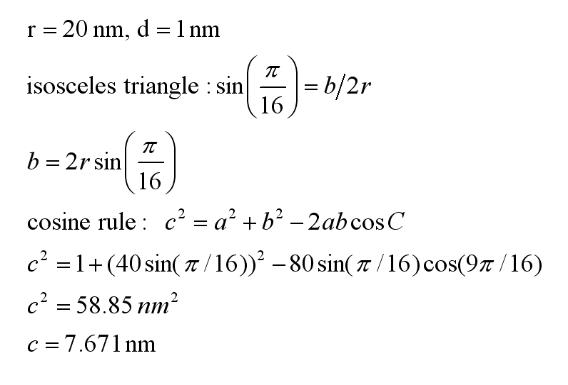
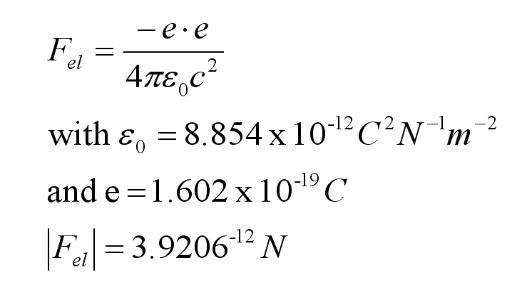

Torque and Drag and Time Reversibility
What we are missing is the drag force exerted by the external water on the rotating flagellum.
There may also be significant internal resistance. Bacteria are tiny and their flagellum, about
6 micrometres long when coiled (the length is variable but our value is reasonable) consists of
a filament 12 nm in radius, but which coils into a helix with a radius of about 200 nm. For such
a small cylinder, even one rotating so fast, the Reynold's number (Re) is estimated to be
much less than one. In swimming bacteria we are dealing with creeping flow, that is the water
behaves as a highly viscous, treacle-like fluid. This poses special problems for locomotion.
Simply trying to swim by waving a paddle from side-to-side (in the manner of some larger
eukaryotic undulipodia) will not effect movement. This is because viscosity dominates at such
low Re and fluid does not drift much in-between motions, rather it behaves more like treacle
and simply pushing a paddle to the left and then to the right, returns the same fluid elements
back where they started and no net movement is achieved: playing the motion in reverse
reveals no change in the fluid. To move through the fluid we need a motion which is not
time-reversible in this way. A corkscrew breaks symmetry in this way. To uncork a bottle one
must rotate the corkscrew in the correct sense, rotating in the opposite sense will not work!
The motion of a corkscrew through a medium is not time-reversible. Bacteria rely on similar
time-symmetry breaking mechanisms. The flagellum is a helix with a definite handedness, like
a corkscrew, and it effectively drills out a core of water as it rotates. Displacement of this core
of water to the rear of the cell drives it forwards by conservation of linear momentum. Thus,
the load on the flagellum could be modeled as the mass of this core of water drilled out from
the water, however, there should also be external friction or viscous drag between this 200
nm diameter cylinder and the surrounding water.
First of all we calculate the torque acting on this 200 nm flagella 'cylinder'.
What we are missing is the drag force exerted by the external water on the rotating flagellum.
There may also be significant internal resistance. Bacteria are tiny and their flagellum, about
6 micrometres long when coiled (the length is variable but our value is reasonable) consists of
a filament 12 nm in radius, but which coils into a helix with a radius of about 200 nm. For such
a small cylinder, even one rotating so fast, the Reynold's number (Re) is estimated to be
much less than one. In swimming bacteria we are dealing with creeping flow, that is the water
behaves as a highly viscous, treacle-like fluid. This poses special problems for locomotion.
Simply trying to swim by waving a paddle from side-to-side (in the manner of some larger
eukaryotic undulipodia) will not effect movement. This is because viscosity dominates at such
low Re and fluid does not drift much in-between motions, rather it behaves more like treacle
and simply pushing a paddle to the left and then to the right, returns the same fluid elements
back where they started and no net movement is achieved: playing the motion in reverse
reveals no change in the fluid. To move through the fluid we need a motion which is not
time-reversible in this way. A corkscrew breaks symmetry in this way. To uncork a bottle one
must rotate the corkscrew in the correct sense, rotating in the opposite sense will not work!
The motion of a corkscrew through a medium is not time-reversible. Bacteria rely on similar
time-symmetry breaking mechanisms. The flagellum is a helix with a definite handedness, like
a corkscrew, and it effectively drills out a core of water as it rotates. Displacement of this core
of water to the rear of the cell drives it forwards by conservation of linear momentum. Thus,
the load on the flagellum could be modeled as the mass of this core of water drilled out from
the water, however, there should also be external friction or viscous drag between this 200
nm diameter cylinder and the surrounding water.
First of all we calculate the torque acting on this 200 nm flagella 'cylinder'.
This torque of about 24 000 pN nm compares to experimentally obtained estimates of about
1500 to 6000 pN nm (e.g. see Inoue et al. 2008. J. Mol. Biol. 376: 1251–1259) and so is about
4 to 16 times too high. However, there are modifications we can make to our model, such as
when calculating the electrostatic force we can assume screening of the charges due to the
surrounding protein of the Mot channel. Proteins typically have dielectric constants of about 5
which would reduce the force and torque 5-fold, bringing it within the experimental value! (We
can also incorporate some repulsion due to the more remote like charges on the rotor ring).
With a dielectric constant of 5 our model torque becomes: 4894 pN nm
First we estimate the Reynold's number (Re) for our rotating flagella cylinder.
1500 to 6000 pN nm (e.g. see Inoue et al. 2008. J. Mol. Biol. 376: 1251–1259) and so is about
4 to 16 times too high. However, there are modifications we can make to our model, such as
when calculating the electrostatic force we can assume screening of the charges due to the
surrounding protein of the Mot channel. Proteins typically have dielectric constants of about 5
which would reduce the force and torque 5-fold, bringing it within the experimental value! (We
can also incorporate some repulsion due to the more remote like charges on the rotor ring).
With a dielectric constant of 5 our model torque becomes: 4894 pN nm
First we estimate the Reynold's number (Re) for our rotating flagella cylinder.


Figure 11. Calculation of Re.
For every torque there is an equal and opposite torque (Newton's Third law) and so if the rotor
drives the flagellum to rotate CCW (counterclockwise) then an equal torque rotates the cell
body CW. In order to test our model it is easier to model drag on the cell body since we can
model this as a one micrometre diameter sphere (or we could model it as a simple rotating
cylinder).
For every torque there is an equal and opposite torque (Newton's Third law) and so if the rotor
drives the flagellum to rotate CCW (counterclockwise) then an equal torque rotates the cell
body CW. In order to test our model it is easier to model drag on the cell body since we can
model this as a one micrometre diameter sphere (or we could model it as a simple rotating
cylinder).








